Background
An ongoing, rapid transition is taking place in molecular biology. In the last 50 years this field was guided by two central dogmas. The first linked the biological function of the protein to its unique three-dimensional structure. The second assumed that specific interactions of proteins are unambiguous. A series of ample experimental evidence demonstrates however, that these concepts need to be revisited. A ‘new view’ is being developed, where conformational diversity of proteins is recognized and furthermore, is required for a variety of functions1. Structural heterogeneity ranges from changes in side-chain conformations to alternative packing of the whole protein chain 2. Pertinent examples are the intrinsically disordered (ID) proteins, which do not adopt a well-defined three-dimensional structure, but may possess transiently structured elements3,4. The discovery of ID proteins caused an earthquake in structural biology and opened novel perspectives. Probably the most exiting one is how conformational heterogeneity promotes multiple functions of the same protein and how these functions can be modulated5.
Monika Fuxreiter has been working on the molecular basis and mechanisms of ID functions6-16. She showed that ID proteins have modular organization with short segments distinguished for interactions6. The characteristic amino acid composition of these motifs can be identified from the primary sequence7. Upon binding their partners, ID regions can fold and adopt a well-defined three-dimensional structure17. In many complexes however, ID regions retain their conformational freedom in the bound form and ambiguous interactions mediated by the ID region are critical for specificity or affinity12,15. These complexes, which are termed as ‘fuzzy’ complexes12, intimidate both dogmas of molecular biology: they lack a well-defined structure and have ambiguous interactions.
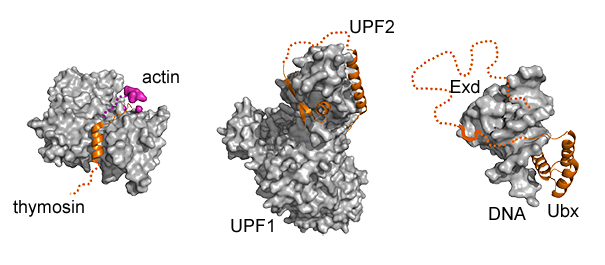
Gallery of fuzzy complexes participating in (from left to right) cytoskeleton formation, nonsense mediated decay and organ development. Bound ID regions are shown by dotted lines.
The idea of fuzziness was proposed by Monika Fuxreiter and Peter Tompa using protein-protein interactions18 . The mechanisms how fuzzy complexes are formed and how they are involved in complex signaling networks are currently explored in our laboratory 19,20.
Fuzzy complexes in cellular networks
We aim to understand how fuzzy complex are exploited for versatile regulatory pathways: i) regulation of protein half-life ii) regulation of cellular location iii) regulation of transcriptional networks. We aim to link fuzzy complexes mediated signaling pathways to disease development.
Evolution and protein dynamics
We investigate how dynamical properties of protein contribute to evolution. i) the role of reorganization energy in enzymatic evolution ii) the role of dynamics in evolution of new protein functions iii) the evolution of disordered protein regions.
References
- James, L.C. & Tawfik, D.S., Conformational diversity and protein evolution–a 60-year-old hypothesis revisited. Trends Biochem Sci 28 (7), 361-368 (2003).
- Dyson, H.J. & Wright, P.E., Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol 6 (3), 197-208 (2005).
- Wright, P.E. & Dyson, H.J., Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol 293 (2), 321-331 (1999).
- Romero, P. et al., Thousands of proteins likely to have long disordered regions. Pac. Symp. Biocomputing. 3, 437-448 (1998).
- Boehr, D.D., Nussinov, R., & Wright, P.E., The role of dynamic conformational ensembles in biomolecular recognition. Nat Chem Biol 5 (11), 789-796 (2009).
- Fuxreiter, M., Simon, I., Friedrich, P., & Tompa, P., Preformed structural elements feature in partner recognition by intrinsically unstructured proteins. J Mol Biol 338 (5), 1015-1026 (2004).
- Fuxreiter, M., Tompa, P., & Simon, I., Local structural disorder imparts plasticity on linear motifs. Bioinformatics 23 (8), 950-956 (2007).
- Solt, I., Magyar, C., Simon, I., Tompa, P., & Fuxreiter, M., Phosphorylation-induced transient intrinsic structure in the kinase-inducible domain of CREB facilitates its recognition by the KIX domain of CBP. Proteins 64 (3), 749-757 (2006).
- Toth-Petroczy, A. et al., Malleable machines in transcription regulation: the mediator complex. PLoS Comput Biol 4 (12), e1000243 (2008).
- Toth-Petroczy, A., Simon, I., Fuxreiter, M., & Levy, Y., Disordered tails of homeodomains facilitate DNA recognition by providing a trade-off between folding and specific binding. J Am Chem Soc 131 (42), 15084-15085 (2009).
- Fuxreiter, M. et al., Malleable machines take shape in eukaryotic transcriptional regulation. Nat Chem Biol 4 (12), 728-737 (2008).
- Tompa, P. & Fuxreiter, M., Fuzzy complexes: polymorphism and structural disorder in protein-protein interactions. Trends Biochem Sci 33 (1), 2-8 (2008).
- Fuxreiter, M. & Tompa, P., Fuzzy interactome: the limitations of models in molecular biology. Trends Biochem Sci 34 (1), 3 (2009).
- Tompa, P. et al., Close encounters of the third kind: disordered domains and the interactions of proteins. Bioessays 31 (3), 328-335 (2009).
- Fuxreiter, M., Simon, I., & Bondos, S., Dynamic protein-DNA recognition: beyond what can be seen. Trends Biochem Sci 36 (8), 415-423 (2011).
- Fuxreiter, M. & Tompa, P., Fuzziness: Structural Disorder in Protein Complexes (Landes BioScience/Springer, Austin, New York, 2011).
- Wright, P.E. & Dyson, H.J., Linking folding and binding. Curr Opin Struct Biol 19 (1), 31-38 (2009).
- Monika Fuxreiter and Peter Tompa (Eds) Fuzziness: Structural disorder in protein complexes, Landes Bioscience/Springer, Austin, NY 2011
- M Fuxreiter (2012) Fuzziness: linking regulation to protein dynamics. Mol Biosystems 8, 168-177
- M. Buljan, G. Chalancon, S. Eustermann, G. Wagner, M. Fuxreiter, A. Bateman, M.M. Babu (2012) Tissue-specific splicing of disordered segments that embed binding motifs rewires protein interaction networks. Mol Cell 46, 871-883.