Structures
|
|
|
p27 |
Ubx | UPF2 |
|
|
|
MBP |
PC4 |
Sic1 |
|
|
|
Ets1 |
HMGB1 |
Oct-1 |
|
||
WH2 domains |
Description of complexes
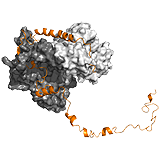
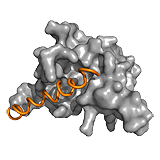
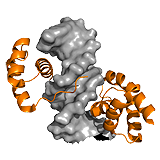
p27Kip1 The p27Kip1 regulates eukaryotic cell division by interacting with cyclin- dependent kinases. In the Cdk2-cyclin complex, the p27 kinase inhibitory domain (KID) blocks the active site of the kinase. The dynamics of the interacting 310 helix segment however, allows the exclusion of Y88 from the active site, which becomes available for phosphorylation by a nontyrosine-receptor kinase. Due to the fuzziness of the C-terminal region, this segment flaps back onto the active site of Cdk2, and gets phosphorylated at T187 via a unimolecular mechanism. This phosphorylation event faciliates interactions with SCF/Skp2 ubiquitin ligase. Ubiquitination at the C-terminal region induces the degradation of p27 Kip1 and causes the progression of the cell cycle. Hence the regulation by p27 is linked to two competitive mechanisms: i) between Y88 and T187 to bind the active site of Cdk2, and ii) between Cdk2/cyclin and SCF/Skp2 to bind T187. The latter is controlled by phosphorylation. Thus fuzziness in p27Kip1 underlies two opposite activities of the same protein. ->
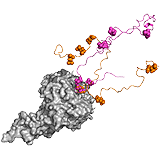
Ubx Hox transcription factors control animal development through region specific differentiation. DNA recognition is carried out by the structured homeodomain (HD), which has high affinity, yet little selectivity in vivo. In the transcription factor Ultrabithorax (Ubx) multiple ID regions control DNA interactions (Figure 1C). The inhibitory I1 and I2 regions reduce affinity of Ubx by 2-fold, and by 40-fold, respectively, whereas the R region improves binding in a length-dependent manner. pH-dependence of DNA binding indicates that I1 directly contacts ionizable residues of the HD binding interface and thus competes with DNA. I2 also acts via competitive mechanisms. On the one hand, I2 sterically blocks DNA contact sites of HD and on the other hand it competes with R for the same transient interactions. Such competitive interplay between I2 and R fine-tunes DNA binding affinity.
Protein–protein interactions or alternative splicing enable ‘context specific gene regulation’ of Ubx; i.e. cognate DNA sequences depend on the cellular context .The I1 region contains a YPWM motif, which mediates communication with protein partners, for example with the Hox co-factor Extradenticle (Exd), which relieves repression of DNA binding by I1. Different Ubx splicing isoforms are produced in a stage- and tissue-specific manner by alternative splicing of three microexons which are located in the region linking the YPWM motif to the HD. Removal of the microexons (alone or in combination) affects Ubx DNA affinity and selectivity both in vitro and in vivo. ->
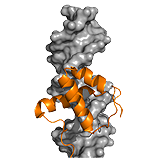
UPF2: The complex of nonsense mediated decay factors UPF1 and UPF2 initiates mRNA degradation. UPF1 and UPF2 interact in a bipartite manner. The C terminal ID region of UPF2 adopts two regular secondary structures upon binding, which are connected by a linker without a rendered structure (poor electron density). This conforms to a clamp-like fuzzy model. The isolated α-helical segment of UPF2 has weaker affinity than the β-hairpin region, the cooperativity between the two segments however, improves binding by 80-fold. Tethering by the linker region is not only beneficial for affinity, but also contributes to organization of the complex on a larger scale. ->
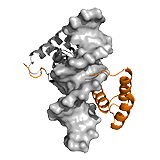
MBP The myelin basic protein (MBP) is part of the mammalian central nervous system that plays a role in maintaining the stability and integrity of the myelin sheath. Myelin signaling is achieved via direct interactions with a variety of molecules, such as calmodulin, actin and tubulin. The proline-rich segment of the membrane bound MBP interacts with diverse SH3 domain-containing ligands (e.g. Yes1, PSD95, cortactin, PexD, Abl, Fyn, c-Src, Itk ). The SH3 domains are typical components of intracellular signaling proteins, including nonreceptor tyrosine kinases, out of which Fyn also influences MBP expression. Besides the direct contacts with the proline residues, formation of all these complexes are facilitated by long-range electrostatic interactions between the ID region of MBP outside the binding context. These interactions in combination with post-translational modifications and alternative splicing contribute to multifunctionality of MBP, and enable to integrate diverse signals from a variety of signaling pathways. ->
PC4 The human positive cofactor 4 (PC4) is a DNA-binding protein that is involved in transcriptional regulation by recruiting general transcription factors to the preinitiation complex. Activity of PC4 is strongly influenced by two ID segments: a serine- and acidic rich (SEAC) and a lysine-rich region, both located at the N-terminal domain (NTD). Although the NTD itself does not have a measurable affinity for DNA, it decreases binding affinity of the C-terminal region of PC4 for ssDNA and reduces DNA unwinding activity of PC4. This is due to transient intra-molecular interactions between the NTD and the structured C-terminal domain (CTD), which compete with DNA binding. Phosphorylation of the lysine-rich reason of the NTD strengthens these interactions, hence reduces binding affinity. PC4 also interacts with various trans-activator domains, for example that of the herpes simplex virion protein 16 (VP16). Contacts with VP16 are also mediated by the disordered NTD, mostly via electrostatic attraction. The opposite effects of the NTD on the activator and DNA binding of PC4 indicates an overlap with the transient intra-molecular contacts of this region with CTD. Thus binding of an external partner interferes with the regulatory potential of this ID region. ->
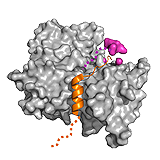
Sic1 The complex of the cyclin-dependent kinase (CDK) inhibitor Sic1 and the SCF ubiquitin ligase subunit Cdc4 is formed in a phosphorylation dependent manner. Sic1 is a multivalent ligand comprising 9 phosphorylation sites, which target only a single binding site on Cdc4. Nevertheless, multiple phosphorylation (at least 6) is required for binding and subsequent degradation of Sic1. Phosphorylation causes only transient, local ordering around the binding motifs, while the whole complex remains dynamic. This enables an interchange between phosphorylation sites, which are in dynamic equilibrium with each other. The degree of phosphorylation fine-tunes the strength of interaction in the complex via long-range electrostatic interactions 62. Hence multiple phosphorylation ensue ultra-sensitivity of the Sic1-Cdc4 binding and ultimately, the cell cycle. ->
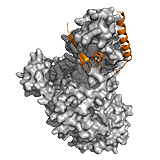
Ets1 The Ets-1 transcription factor regulates various genes, and is involved in stem cell development and tumorigenesis. Ets-1–DNA binding is regulated by an autoinhibitory region and requires the HI-1 helix to unfold. Interactions with DNA are further attenuated by a serine-rich region (SRR), which is disordered in both the free and complex form. Phosphorylation of five sites within the SRR region gradually reduces binding affinity up to ~1000 fold. Although phosphorylation itself has minimal impact on the secondary structure of SRR, it interferes with the formation of transient intraprotein contacts. The most pronounced differences are in the dynamic properties of the HI-1 autoinhibitory helix and recognition helices H1 and H3. These units form a hydrophobic network, whose motions are dampened by SRR phosphorylation. Truncating the SRR region gradually increases the mobility of these residues and facilitates HI-1 unfolding, a process that is required for DNA binding. Thus the distant ID region in Ets-1 perturbs the dynamics of the protein-DNA interface and modulates the conformational transition leading to the tight, specific complex.
In both examples, phosphorylation conforms to the ‘incremental rheostat’ type regulatory mechanism, which was originally proposed for Ets-1. In this model, gradual changes in DNA binding affinity take place via multiple phosphorylation events, in contrast to a phosphorylation-dependent on/off switch mechanism. The ‘incremental rheostat’ regulation provides a sensitive mechanism by which to control transcription in response to different environmental signals (e.g. Ca2+ signaling).
Alternative splicing of ETS1 removes the entire disordered SRR region; phosphorylation of this region reduces affinity for DNA via modulating the flexibility of the interface. Hence, the activity of human Ets-1 is differentially regulated by two distinct mechanisms: phosphorylation and alternative splicing. ->
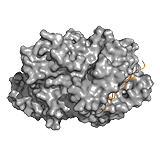
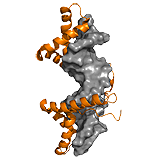
HMGB1 A competitive mechanism is observed for the high mobility group (HMG) protein B1. HMGB1 participates in various nuclear processes by contacting its target DNA via two HMG boxes, a process which is negatively regulated by the disordered C-terminal tail. The tail stabilizes HMGB1, but does not perturb its secondary or tertiary structure. In the absence of DNA, the two HMG boxes assemble on the acidic C-terminal tail and the binding surfaces are only transiently exposed. The tail-bound collapsed form is in dynamic equilibrium with an extended DNA binding competent form, in which the tail remains disordered. Because the tail screens interactions between the two HMG boxes, it affects DNA recognition in a length-dependent manner. ->
Oct-1 The separate DNA-binding motifs in multidomain proteins are often connected by highly flexible/ID linkers, which facilitate the target search along the DNA. The linkers are usually absent from the crystal structures of complexes and thus preserve their conformational heterogeneity, when bound to DNA.
Pit-1, Oct-1, Unc-86 (POU) domain transcription factors recognize different bipartite DNA motifs depending on the length and sequence of the connecting region. The linker of the octamer binding factor 1 (Oct-1) connects two helix-turn-helix motifs. It cannot be seen in the complex and is also sensitive to proteolysis. Shortening this segment or modifying its amino acid composition cause dramatic decreases in binding to either sites. Charge distribution of the linker, with an essential negatively charged Glu, is critical for DNA recognition. Indeed, this region does not merely contribute to electrostatic attractions. ->
WH2 domains Alternative contacts may impart different morphology on larger assemblies. For example, actin polymerization is regulated by various proteins containing tandem repeats of WH2 domains (eg. thymosine, ciboulot, Spire, Cordon bleu) 25. Binding of the regulators to the disordered subdomain-2 of actin induces their folding. The variable interactions of the WH2 C-terminal tail affects the spacing between the ordered parts of the different WH2 domains and thereby the organisation of the actin polymer.
The degree of dynamics of the C-terminal tail of thymosine, which remains disordered even when bound to actin, has a significant impact on actin polymerization. An electrostatic interactions between an ionic pair stabilizes the WH2-actin complex, the magnitude of which depends on the ionic strength. At low ionic strength, a strong interaction is formed and dissociation of WH2 tail from actin is unflavored. In contrast, at high ionic strength the tail remains highly dynamic, and easily disassembles from actin. This facilitates actin polymerization at high ionic strength and sequestration of the polymer at low ionic strength. ->